serhii.net
In the middle of the desert you can say anything you want
-
Day 1731 (28 Sep 2023)
LM Benchmarks notes
Context: 230928-1527 Evaluation benchmark for DE-UA text Here I’ll keep random interesting benchmarks I find.
HELM
- Code
- Random code sample from all common-sense scenarios: helm/src/helm/benchmark/scenarios/commonsense_scenario.py at main · stanford-crfm/helm
- Scenarios: Holistic Evaluation of Language Models (HELM)
GLUECoS
code: GLUECoS/Code at master · microsoft/GLUECoS
Cross-lingual
XGLUE
- microsoft/XGLUE: Cross-lingual GLUE
- Interesting enough for list of tasks and code
- Data: XGLUE
Other
- How truthful is GPT-3? A benchmark for language models — LessWrong
- [2210.14986] Large language models are not zero-shot communicators
- LMentry!
- https://github.com/aviaefrat/lmentry
- With regex-based evaluations for the tasks!
BIGBench
Leaderboards
’evaluation harness’es
- lm-evaluation-harness/lm_eval/tasks at big-refactor · EleutherAI/lm-evaluation-harness - very clean code etc., currently the branch big-rewrite is the best one
- ParlAI/parlai/tasks/multinli/test/multinli_test.yml at main · facebookresearch/ParlAI
- openai/evals: Evals is a framework for evaluating LLMs and LLM systems, and an open-source registry of benchmarks.
- TheoremOne/llm-benchmarker-suite: LLM Evals Leaderboard
- the most meta of all similar suites I’ve seen
Random relevant code
Ideas for Ukrainian LM eval tasks
Context: 230928-1527 Evaluation benchmark for DE-UA text
Shortlist
Tasks
- 231024-1704 Master thesis task CBT
- the three existing
ua_datasetstask - RU/UA Interference
- Фемінітиви - autocompleting recent language
- WSD
How
- (partially auto-generated) Google Spreadsheet for anything requiring manual changes/creation
Ideas / general
- UCL-DARK/ludwig · Datasets at Hugging Face has an example of 0-shot, 1-shot etc. added to the dataset itself, as folds!
General
- would be cool to create ones from different HELM scenarios1
- would be cool to find not-work-intensive ways to create this data coming from other benchmarks (e.g. classify headers by article tags etc.)
- especially find cool ways to use the annotated corpora I found
- I could generate my own tasks easily with some kind of annotation tool, a la label-parser, and annotate bits of Ukrainian Wikipedia2 a la SQuAD
- I could use some google translate API3 thing
- and manually check the translations! AND HAVE TWO DIFFERENT DATASETS AND DO GRAPHS OF DIFFERENCES/CHANGES!!!
- LLMs shouldn’t scare me from including easy tasks - smaller LMs exist in many contexts!
- For simplicity and ease of inclusion to other benchmarks, I shouldn’t do anything requiring too much code. Maybe even literally limit myself to exact match or multiple-choice questions, along with prompts or something, so that the HF datasets are enough.
- And for simplicity in uploading the datasets to HF
Ideas
- Based on LinguisticAndInformationSystems/mphdict: Digital lexicographic systems Ukrainian language + (the grammatical dictionary, synonymous dictionary, etymological dictionary +):
- Find the best synonym for $word
- Tasks on Ukrainian/Russians verbs of motion4:
- Correct verb of motion for hypothetical situations
- Ask whether certain words rhyme
- especially ones where the letter make it seem like they do, but they don’t
- ask for correct stressing of individual words?5
- Чи правильно використані фразеологізми
- Find the correct tag for the title of an article, from the possible parallel corpus: 231002-2311 230928-1651 Random stuff about the Masterarbeit#UA-RU parallel corpus
- Children’s book test<
@taskCBT(2015) z/d/>- Gutenberg has no Ukrainian books, but Anna’s archive does and many of them are actually stories and epub/fb2: казки - Search - Anna’s Archive
- One could filter them by decade etc. for copyright
- Then POS-tag, and automatically generate examples
- Yes/no questions:BoolQ: Exploring the Surprising Difficulty of Natural Yes/No Questions - ACL Anthology
- Russian-language interference!
- Remember how a number of “ukrainian” datasets of HF hub are actually Russian
- Resources:
- СЛОВОВЖИВАННЯ | Горох — українські словники
- відноситися - Антисуржик. Словник «українського» суржика
- Словник-антисуржик онлайн
- Антисуржик (словник) - Русский/украинский язык, культура - Форум Днепродзержинск-Каменское
EXCELLENT! Мова – не калька: словник української мови - Тарас Береза - Тека авторів - Чтиво- parse -> estimate frequency -> include only the most frequent?
- A lot of the examples there are let’s say questionable to my central-Ukrainian ear
- голий -> “У костюмі (в одежі) Адама і Єви; у чому мати [на світ] народила.” alrighty then
- Льотчик -> летун
- Ліберія -> “Вільна країна” I’m done
- I want RU interference (!= суржик); I want RU interference (!= стилістика)
- Some kind of filtering is definitely needed. Could be as easy as putting “1” in rows of a spreadsheet
- https://chtyvo.org.ua/authors/Tykhyi_Oleksii/Slovnyk_movnykh_pokruchiv.pdf
- Суржиково-український словник
- has really nice intro!
- Українське життя в Севастополi Юрій Гнаткевич СЛОВНИК-АНТИСУРЖИК ^ff5ccc
- СЛОВОВЖИВАННЯ | Горох — українські словники
- Frame as multiple-choice task! Or boolean? Or “Is this a correct sentence”?
- I really like this: `“Цей студент [взявся за/почав] дослідження важкої теми.”
- For fun, here’s ChatGPT lying about prefixes: https://chat.openai.com/share/0eda9061-d2cf-46bc-ad45-38cc6e58934a
- False friends!
- Here’s an itemized list: Фальшиві друзі перекладача — Вікіпедія
- сир/сыр, неділя/неделя/…
- Here’s an itemized list: Фальшиві друзі перекладача — Вікіпедія
- ChatGPT ideas:
-
On the semantic front, exploit polysemy and homonymy differences. Formulate sentences with words that have multiple meanings in Russian, but those meanings have distinct equivalents in Ukrainian. This will challenge the model to accurately discern the intended sense based on context.
-
- Implicature6:
- LMEntry-lite-UA7
- Subset of the LMentry questions, translated to UA, with exact matches
- will do this! here 231203-1745 Masterarbeit eval task LMentry-static-UA
- Good old fashioned perplexity. Getting a Ukrainian reference corpus a la Wikipedia and benchmarking on it was always allowed
- Or Telegram, or news comments!
- Look into stability of models to OCR errors! Either scan some old Ukrainian book I have or simulate OCR errors like I did for BxE!
- I’m not the first who thought of this8
- ocr ukrainian - Google Scholar
- Something about the recent changes in UA, both the new 2019 orthography and feminitives 9 is now here: 231204-1642 Masterarbeit evaluation task new UA grammar and feminitives
- Use the UPravda dataset, replace bits with synonyms to get around contamination, and then do classification / entailment /…!!!
- Use the bold bits ‘дослівно’ etc., and match the пряма мова to the correct article title/text?Залужний востаннє поговорив з Міллі на його посаді | Українська правда
Neat datasets
- I could use these open data for petitions to match the text by type! 2.37. Дані про електронні петиції Вінницької міської територіальної громади, у тому числі, осіб, що їх підписали, та результати розгляду - Петиції 2020 - OpenData.gov.ua
Work required
Babi
From 10, automatically generated!
- facebookarchive/bAbI-tasks at ccd8fd6af76d35346109e7bb5f7de9138d055e01
- bAbI-tasks/lua/babi/World.lua at ccd8fd6af76d35346109e7bb5f7de9138d055e01 · facebookarchive/bAbI-tasks
- !!! bAbI-tasks/lua/babi/tasks/worlds/world_basic.txt at ccd8fd6af76d35346109e7bb5f7de9138d055e01 · facebookarchive/bAbI-tasks
I could also use a graph-based approach? As in create an ontology, ask questions about it?..
Or split it into multiple sub-tasks! one for time, one for y/n, etc.?
Make my own IMDB dataset
Find
some popular website with comments and ratings, do sentiment analysis: can I scrape https://rozetka.com.ua/jagermeister_4067700015532_/p4971091/comments/ ?- Also: comfy.ua: Відгуки про Pecham Professional 600 мл черный)
- Someone did something similar! vkovenko/cross_domain_uk_reviews · Datasets at Hugging Face
Not all comments are in UA but I can filter it.
Use movie subtitles for basic dialogs
- e.g. Top rated movies - opensubtitles.com | opensubtitles.com
- OpenSubtitles Dataset | Papers With Code
- but then to do what?..
Literally google-translate other benchmarks and see what happens
- E.g. this is nice: How truthful is GPT-3? A benchmark for language models — LessWrong
Where to get ideas
- the list of tasks/areas in Natural Language Processing | Papers With Code is another source of inspiration
- Read a UA/RU language textbook for other cool hard things like the verbs of motion
- Глянути завдання ЗНО!
Where to get data
- Ask people I know for non-classified documents from their work that aren’t googleable! And measure e.g. perplexity on it, and add a canary to it when uploading the benchmark itself!
- (or upload it as huggingface dataset so it’s not indexed in my github repo)
- (or upload it encrypted as ceasar cypher and include the python script or
cat task_text.txt | rot13or whatever)
Existing tasks
UA-only
From fido-ai/ua-datasets: A collection of datasets for Ukrainian language:
Multilingual including UA
- Belebele Dataset | Papers With Code is a " multiple-choice machine reading comprehension (MRC) dataset", 122 languages
- facebookresearch/belebele: Repo for the Belebele dataset, a massively multilingual reading comprehension dataset.
- facebook/flores · Datasets at Hugging Face has literally an example in Ukrainian <3
- I CAN USE IT AS SENTENCE CLASSIFICATION BENCHMARK!!!!
- SIB-200: A Simple, Inclusive, and Big Evaluation Dataset for Topic Classification in 200+ Languages and Dialects | Papers With Code
- KGQA/QALD_9_plus: QALD-9-Plus Dataset for Knowledge Graph Question Answering - one of the 9 langs is Ukrainian! One could theoretically convert the entities into text
- One could look for similar datasets over wikimagic in English, then take the name of the corresponding Ukrainian page
- Training prompts for instruction finatuning, translated to UA too, can be used for matching?.. MBZUAI/Bactrian-X · Datasets at Hugging Face
- Measuring knowledge retrieval from LMs in many languages: https://huggingface.co/datasets/Polyglot-or-Not/Fact-Completion / daniel-furman/polyglot-or-not: [arXiv pre-print] Are foundation language models multilingual knowledge bases?
- Actually quite cool! Multilingual leaderboard and all that
- Can be neatly compared to a pure-UA model
- Pretty much everything mentioned in the chatGPT-beyond-English paper<
@laiChatGPTEnglishComprehensive2023ChatGPT Beyond English (2023) z/d/>
Random
This is a dictionary that has homonyms as column in the CSV: tamila-krashtan/UkrEtymDict: Revised database of Ukrainian Etymological Dictionary
-
ParlAI/parlai/tasks/squad2/test/squad2_index_test.yml at main · facebookresearch/ParlAI ↩︎
-
matheuss/google-translate-api: A free and unlimited API for Google Translate :dollar::no_entry_sign: ↩︎
-
lang-uk/ukrainian-word-stress-dictionary: Dictionary of word stresses in the Ukrainian language 🇺🇦 ↩︎
-
<_(@Todorov2022) “An Assessment of the Impact of OCR Noise on Language Models” (2022) / Konstantin Todorov, Giovanni Colavizza: z / / _> ↩︎
-
<_(@synchak2023feminine) “Feminine personal nouns in ukrainian: Dynamics in a corpus” (2023) / Vasyl Starkoand Olena Synchak: z / / _> ↩︎
-
Babi: <
@westonAICompleteQuestionAnswering2015Towards AI-Complete Question Answering (2015) z/d/> / Holistic Evaluation of Language Models (HELM) ↩︎
- Code
-
Day 1722 (19 Sep 2023)
seaborn label bars in histogram plot
The ’new’ function in matplotlib for this is matplotlib.pyplot.bar_label — Matplotlib 3.8.0 documentation (ty Revisions to Display count on top of seaborn barplot [duplicate] - Stack Overflow):
ax = sns.histplot(df.langs_parsed) #ax.set_xlabel("Language(s)") #ax.set_ylabel("# of files") for i in ax.axes.containers: ax.bar_label( i, )The second link has infos about barplot, catplot, and countplot too!
If the text goes over the limit and the light-gray background of seaborn’s theme or something, increase the limit as:
ylim = ax.axes.get_ylim()[1] new_ylim = ylim + 300 ax.axes.set_ylim(0, new_ylim) # you can also set padding of the labels in px and Text (https://matplotlib.org/stable/api/text_api.html#matplotlib.text.Text) properties: for ax in g.axes.containers: g.bar_label(ax, padding=-10,fontsize=5)Disabling scientific notation / setting format
EDIT 2023-10-06: To disable scientific notation, one can use the
fmt=argument (seebar_labeldocu) where one can pass a format, including as f-string:for i in ax.axes.containers: ans = ax.bar_label( i, fmt="{:,.2f}", )There’s also a parameter that decides at which point to start to use sci. notation, I think I closed the tab with the link though+
-
Day 1717 (14 Sep 2023)
German NLP resources
It includes a really cool list of corpora!
And at the end has a list of other such pages for other languages etc.
Also: deutschland · PyPI: “A python package that gives you easy access to the most valuable datasets of Germany.”
-
Day 1714 (11 Sep 2023)
Latex print or not the entire bibliography from a file
The LREC Author’s Kit prints all things in the .bib file and it uses
\nocite{*}for that.The Internet from 2009 agreess that’s the way to go : Biblatex - Printing all entries in .bib file (cited and not)
Removing this line removes the printout.
Lastly, the link above shows printing separate bibliographies; the LREC Author’s kit does something different for the same:
\subsection{Language Resource References} Language resource references should be listed in alphabetical order at the end of the paper. \nocite{*} \section{Bibliographical References}\label{sec:reference} \bibliographystyle{lrec-coling2024-natbib} \bibliography{lrec-coling2024-example} \section{Language Resource References} \label{lr:ref} \bibliographystylelanguageresource{lrec-coling2024-natbib} \bibliographylanguageresource{languageresource}
-
Day 1711 (08 Sep 2023)
Latex page-breaks
TL;DR
\newpage~\newpage~\newpage~\newpagefor 3 empty pages\newpagedoesn’t always work for well me in, esp. not in the IEEE and LREC templates. Either only one column is cleared, or there are issues with images/tables/… positions.\clearpageworks for me in all cases I’ve tried.EDIT: but only one page, not multiple! For multiple empty pages one after the other this1 does the trick:
\newpage ~\newpageChatGPT thinks it works because
~being a non-breaking space makes LaTex try to add both empty pages on the same page, leading to two empty pages. Somehow allowing a newline between new pages makes it interpret both pages as the same command, since it’s already a new page.
-
Day 1639 (28 Jun 2023)
Everything I know about saving plots in matplotlib, seaborn, plotly, as PNG and vector PDF/EPS etc.
Seaborn saving with correct border
When saving seaborn images there was weirdness going on, with borders either cutting labels or benig too big.
Solution:
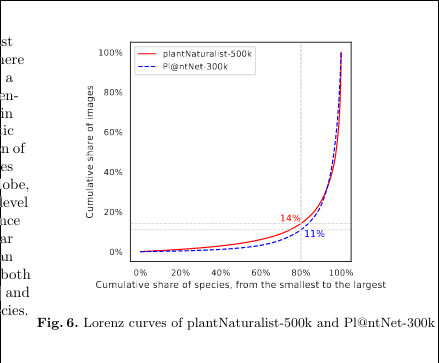
# bad: cut corners ax.figure.savefig("inat_pnet_lorenz.png") # good: no cut corners and OK bounding box ax.figure.savefig("inat_pnet_lorenz.png", bbox_inches="tight")Save as PDF/EPS for better picture quality in papers
EDIT 2023-12-14
Paper reviewer suggested exporting in PDF, which led me to graphics - Good quality images in pdflatex - TeX - LaTeX Stack Exchange:
Both gnuplot and matplotlib can export to vector graphics; file formats for vector graphics are e.g. eps or pdf or svg (there are many more). As you are using pdfLaTeX, you should choose pdf as output format, because it will be easy to include in your document using the graphicx package and the \includegraphics{} command.
Awesome! So I can save to PDF and then include using the usual code (edit - eps works as well). Wow!
Plotly
Static image export in Python:
fig.write_image("images/fig1.png")PDF works as-is as well, EPS needs the poppler library but then works the same way
For excessive margins in the output PDFs:]
fig.update_layout( margin=dict(l=20, r=20, t=20, b=20), )Overleaf antialiasing blurry when viewing
When including a PDF plot, I get this sometimes:

This is a problem only when viewing the PDF inside qutebrowser/Overleaf, in a normal PDF viewer it’s fine!
-
Day 1633 (22 Jun 2023)
Vaex iterate through groups
Didn’t find this in the documentation, but:
gg = ds.groupby(by=["species"]) lg = next(gg.groups) # lg is the group name tuple (in this case of one string) group_df = gg.get_group(lg)
-
Day 1632 (21 Jun 2023)
Zotero web version for better tabs + split view
Web zotero
Looking for a way to have vertical/tree tabs, I found a mention of the zotero web version being really good.
Then you can have multiple papers open (with all annotations etc.) in different browser tabs that can be easily navigated using whatever standard thing one uses.
You can read annotations but not edit them. Quite useful nonetheless!
Split view
PDF reader feature request: open the same pdf twice in split screen - Zotero Forums: View -> Split Horizontally/Vertically!
It’s especially nice for looking at citations in parallel to the text.
Overleaf bits
Shortcuts
vim!
EDIT 2023-12-05 Overleaf has Vim bindings! Enable-able in the project menu. There are unofficially supported ways to even make custom bindings through TamperMonkey
Shortcuts
Kurz und gut
- Ctrl+Enter compiles the project
- Bold/italic work as expected,
<C-b/i>. Same for copypaste etc. - Advanced reference search: is cool.
- Comments:
<C-/>for adding%-style LaTex comments.<C-S-c>for adding Overleaf comments
Bible
Overleaf Keyboard Shortcuts - Overleaf, Online LaTeX Editor helpfully links to a PDF, screenshots here:
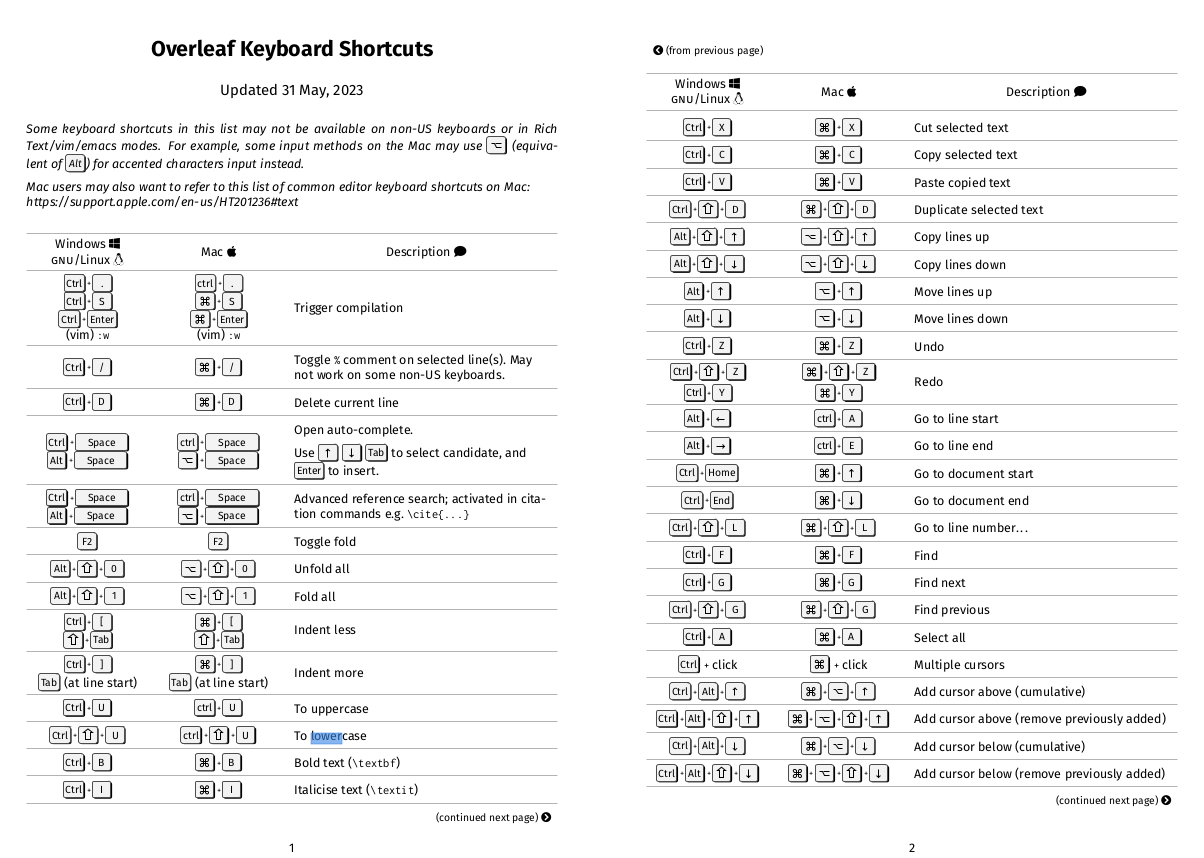
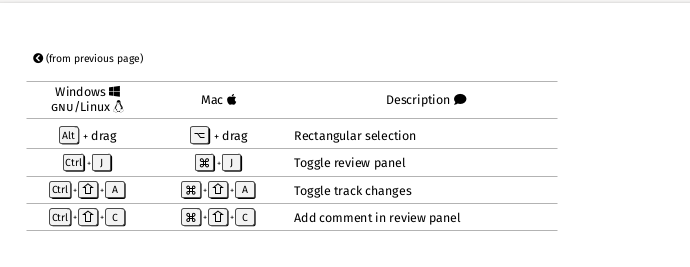
It seems to have cool multi-cursor functionality that might be worth learning sometime.
Templates
Overleaf has a lot of templates: Templates - Journals, CVs, Presentations, Reports and More - Overleaf, Online LaTeX Editor
If your conference’s is missing but it sends you a .zip, you can literally import it as-is in Overleaf, without even unpacking. Then you can “copy” it to somewhere else and start writing your paper.
Bits and pieces
- Renaming the main file to sth like
0paper.texmakes it appear on top, easier to find.